Note
Click here to download the full example code
Synthetic fMRI data example¶
Example to recover the different neural temporal activities, the associated functional networks maps and the HRFs per ROIs in the fMRI data, on simulated fMRI data.
Contents
# Authors: Hamza Cherkaoui <hamza.cherkaoui@inria.fr>
# License: BSD (3-clause)
import os
import numpy as np
import matplotlib.pyplot as plt
from hemolearn.simulated_data import simulated_data
from hemolearn import deconvolution
Create plotting directory¶
plot_dir = 'plots'
if not os.path.exists(plot_dir):
os.makedirs(plot_dir)
Generate the synthetic data¶
TR = 1.0
n_voxels, n_atoms, n_times_valid, n_times_atom, snr = 100, 2, 200, 30, 1.0
noisy_X, _, u, v, z, hrf_rois = simulated_data(n_voxels=n_voxels,
n_times_valid=n_times_valid,
n_times_atom=n_times_atom,
snr=snr)
Distangle the neurovascular coupling from the neural activation¶
results = deconvolution.multi_runs_blind_deconvolution_multiple_subjects(
noisy_X, t_r=TR, hrf_rois=hrf_rois, n_atoms=n_atoms,
deactivate_v_learning=True, prox_u='l1-positive-simplex',
n_times_atom=n_times_atom, hrf_model='scaled_hrf',
lbda_strategy='ratio', lbda=0.5,
u_init_type='gaussian_noise', max_iter=30, get_obj=1,
raise_on_increase=True, random_seed=None,
n_jobs=4, nb_fit_try=4, verbose=0)
z_hat, _, u_hat, a_hat, v_hat, v_init, _, pobj, times = results
z_hat, u_hat, a_hat, v_hat = z_hat[0], u_hat[0], a_hat[0], v_hat[0]
Re-label the components¶
u_0_true = u[0, :]
u_1_true = u[1, :]
z_0_true = z[0, :].T.ravel()
z_1_true = z[1, :].T.ravel()
u_0_hat = u_hat[0, :]
u_1_hat = u_hat[1, :]
z_0_hat = z_hat[0, :].T.ravel()
z_1_hat = z_hat[1, :].T.ravel()
prod_scal_0 = np.dot(z_0_hat, z_0_true)
prod_scal_1 = np.dot(z_0_hat, z_1_true)
if prod_scal_0 < prod_scal_1:
tmp = z_0_hat
z_0_hat = z_1_hat
z_1_hat = tmp
tmp = u_0_hat
u_0_hat = u_1_hat
u_1_hat = tmp
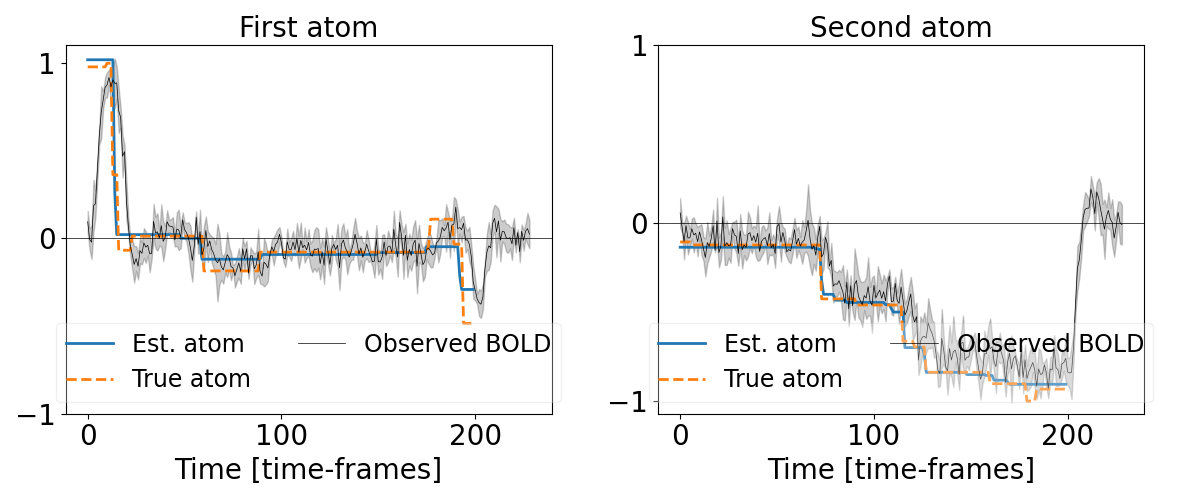
Plot the temporal activations¶
plt.figure("Temporal atoms", figsize=(12, 5))
plt.subplot(121)
plt.plot(z_0_hat, lw=2.0, label="Est. atom")
plt.plot(z_0_true, linestyle='--', lw=2.0, label="True atom")
x_0 = noisy_X[np.where(u_0_true > 0)[0], :]
x_0 /= np.repeat(np.max(np.abs(x_0), axis=1)[:, None], noisy_X.shape[1], 1)
t = np.arange(noisy_X.shape[1])
mean_0 = np.mean(x_0, axis=0)
std_0 = np.std(x_0, axis=0)
borders_0 = (mean_0 - std_0, mean_0 + std_0)
plt.plot(mean_0, color='k', lw=0.5, label="Observed BOLD")
plt.fill_between(t, borders_0[0], borders_0[1], alpha=0.2, color='k')
plt.axhline(0.0, color='k', linewidth=0.5)
plt.xticks([0, n_times_valid/2.0, n_times_valid], fontsize=20)
plt.yticks([-1, 0, 1], fontsize=20)
plt.xlabel("Time [time-frames]", fontsize=20)
plt.legend(ncol=2, loc='lower center', fontsize=17, framealpha=0.3)
plt.title("First atom", fontsize=20)
plt.subplot(122)
plt.plot(z_1_hat, lw=2.0, label="Est. atom")
plt.plot(z_1_true, linestyle='--', lw=2.0, label="True atom")
x_1 = noisy_X[np.where(u_1_true > 0)[0], :]
x_1 /= np.repeat(np.max(np.abs(x_1), axis=1)[:, None], noisy_X.shape[1], 1)
mean_1 = np.mean(x_1, axis=0)
std_1 = np.std(x_1, axis=0)
borders_1 = (mean_1 - std_1, mean_1 + std_1)
plt.plot(mean_1, color='k', lw=0.5, label="Observed BOLD")
plt.fill_between(t, borders_1[0], borders_1[1], alpha=0.2, color='k')
plt.axhline(0.0, color='k', linewidth=0.5)
plt.xticks([0, n_times_valid/2.0, n_times_valid], fontsize=20)
plt.yticks([-1, 0, 1], fontsize=20)
plt.xlabel("Time [time-frames]", fontsize=20)
plt.legend(ncol=2, loc='lower center', fontsize=17, framealpha=0.3)
plt.title("Second atom", fontsize=20)
plt.tight_layout()
filename = "activations.png"
filename = os.path.join(plot_dir, filename)
plt.savefig(filename, dpi=150)
print("Saving plot under '{0}'".format(filename))
Out:
Saving plot under 'plots/activations.png'
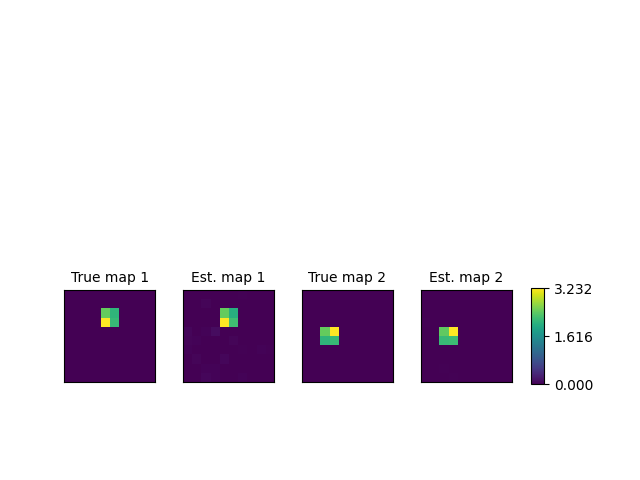
Plot the spatial maps¶
fig, axes = plt.subplots(nrows=1, ncols=4)
len_square = int(np.sqrt(n_voxels))
l_u = [u_0_true.reshape(len_square, len_square),
u_0_hat.reshape(len_square, len_square),
u_1_true.reshape(len_square, len_square),
u_1_hat.reshape(len_square, len_square)]
l_max_u = [np.max(u) for u in l_u]
max_u = np.max(l_max_u)
amax_u = np.argmax(l_max_u)
l_name = ["True map 1", "Est. map 1", "True map 2", "Est. map 2"]
l_im = []
for ax, u, name in zip(axes.flat, l_u, l_name):
l_im.append(ax.matshow(u))
ax.set_title(name, fontsize=10)
ax.set_xticks([])
ax.set_yticks([])
fig.subplots_adjust(bottom=0.1, top=0.5, left=0.1, right=0.8,
wspace=0.3, hspace=0.2)
cbar_ax = fig.add_axes([0.83, 0.2, 0.02, 0.2])
cbar = fig.colorbar(l_im[amax_u], cax=cbar_ax)
cbar.set_ticks(np.linspace(0.0, max_u, 3))
filename = "spatial_maps.png"
filename = os.path.join(plot_dir, filename)
plt.savefig(filename, dpi=150)
print("Saving plot under '{0}'".format(filename))
Out:
Saving plot under 'plots/spatial_maps.png'
Total running time of the script: ( 0 minutes 3.767 seconds)
